> x<-c(3.5, 3.6, 7.8, 9.6, 5.7, 8.9, 6.3)
> y<-c(1.0, 2.7, 8.9, 6.5, 8.9, 6.5,12.5,10.2, 1.2)
> t.test(x,y,alternative=c("two.sided"),var.equal=TRUE,conf.level=0.95)
Two Sample t-test
data: x and y
t = -0.0018, df = 14, p-value = 0.9986
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-3.760075 3.753726
sample estimates:
mean of x mean of y
6.485714 6.488889
Значения
t = -0.0018 (значение критериальной статистики), число степеней свободы равно 14.
p-value = 0.9986, т.е. чтобы отвергнуть гипотезу, нужно допустить 99.86% ошибки.
95% доверительный интервал (-3.760075, 3.753726). Поскольку наше значение в него попадает, то нулевая гипотеза принимается на 5% уровне значимости.
Если равенство дисперсий не проверялось, или гипотеза о равенстве не принимается, то вызов критерия выглядит так:
> t.test(x,y,alternative=c("two.sided"),var.equal=FALSE, conf.level=0.95)
Welch Two Sample t-test
data: x and y
t = -0.0019, df = 13.242, p-value = 0.9985
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-3.545004 3.538655
sample estimates:
mean of x mean of y
6.485714 6.488889
Число степеней свободы теперь 13.242 вместо 14, и границы доверительного интервала несколько изменились.
Приведем пример для проверки нулевой гипотезы о равенстве матожиданий для парной выборки (выборки должны быть одинаковой длина):
> x<-c(3.5, 3.6, 7.8, 9.6, 5.7, 8.9, 6.3, 8.3, 4.5)
> y<-c(1.0, 2.7, 8.9, 6.5, 8.9, 6.5,12.5,10.2, 1.2)
> t.test(x,y,alternative=c("two.sided"),var.equal=TRUE, paired=TRUE)
Paired t-test
data: x and y
t = -0.0202, df = 8, p-value = 0.9843
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-2.554391 2.509946
sample estimates:
mean of the differences
-0.02222222
Значения:
t = -0.0202 (значение критериальной статистики), число степеней свободы равно 8.
p-value = 0.9943, т.е. чтобы отвергнуть гипотезу, нужно допустить 99.86% ошибки.
95% доверительный интервал (-2.554391, 2.509946). Поскольку наше значение в него попадает, то нулевая гипотеза принимается на 5% уровне значимости.
Критерии Бартлетта и Кохрана (Сравнение нескольких дисперсий нормальных генеральных совокупностей по выборкам).
Описание
Критерий Барлетта используется для проверки гипотезу об однородности (равенстве) нескольких дисперсий, полученных по выборкам разного объема. Для этого рассчитывают среднюю арифметическую исправленных дисперсий, взвешенную по числам степеней свободы:
 ,
,
где 
 число степеней свободы для i -й выборки объема
число степеней свободы для i -й выборки объема  ,
,  - выборочная дисперсия дисперсия i -й выборки,
- выборочная дисперсия дисперсия i -й выборки,  - общее число степеней свободы, и
- общее число степеней свободы, и  - число выборок.
- число выборок.
В качестве критериальной статистики для проверки гипотезы об однородности дисперсий используют критерий Бартлетта:
 ,
,
имеющая распределение  , где
, где
 ,
,

Одной из функций, осуществляющей проверку данного критерия в R является bartlett.test()
Описание функции
bartlett.test (x, g...)
Параметры
| x | числовой вектор значений, или список числовых значений векторов, или объекты линейной модели (класса "lm"). |
| g | вектор или фактор, дающий группу для соответствующих элементов x. Игнорируемый, если x - список. |
Примечание
Если x - список, его элементы будут взяты как выборки и g игнорируется, и можно просто использовать bartlett.test(x). Если выборки еще не содержатся в списке, используют bartlett.test(list (x...)).
Критические значения (правосторонний критерий) находятся по таблице распределения 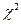 с
с  степенями свободы [2,стр.329] или используют функцию вычисления квантилей распределения Хи-квадрат qchisq(p,df).
степенями свободы [2,стр.329] или используют функцию вычисления квантилей распределения Хи-квадрат qchisq(p,df).
Пример
> x1<-c(3.5, 3.6, 7.8, 9.6, 5.7, 8.9, 6.3)
> x2<-c(1.0, 2.7, 8.9, 6.5, 8.9, 6.5,12.5,10.2, 1.2)
> x3<-c(3.6,7.8,9.6,5.7,8.9)
> x4<-c(2.7,8.9,6.5,8.9)
Дисперсии выборок равны соотвественно 5.86, 16.75, 6.05 и 8.57, нулевая гипотеза H0 –дисперсии всех генеральных совокупностей равны между собой, уровень значимости – 5%.
> bartlett.test(list(x1,x2,x3,x4))
Bartlett test of homogeneity of variances
data: list(x1, x2, x3, x4)
Bartlett's K-squared = 2.2368, df = 3, p-value = 0.5247
Значения
Bartlett's K-squared = 2.2368 (значение критериальной статистики теста Бартлетта), число степеней свободы 3,
p -value = 0.5247, т.е. отвергнуть гипотезу H0 можно только при допустимой ошибке в 52.47%. Следовательно, гипотеза об однородности дисперсий принимается на 5% уровне значимости.
Если объем выборок (примерно) одинаковый, то может использоваться тест экстремальных значений Кохрана (Cochran) из пакета outliers, реализуемый функцией cochran.test().
Описание функции
cochran.test(object,data)
Параметры
| object | числовой вектор, содержащий значения дисперсий для каждой выборки 
|
| data | числовой вектор, содержащий объем каждой выборки |
В в качестве критериальной статистики используется

а для вычисления критических значений – функция вычисления квантилей распределения Кохрана qcochran(p, n, k) из того же пакета, где p - доверительная вероятность, n – объем одной выборки (если объемы различаются, то берется среднее значение), k – число выборок.
Пример
Используем в примере те же выборки, что и в предыдущем случае, объем выборок 7, 9, 5 и 4 элементов соответственно. Нулевая гипотеза H0 – дисперсии всех генеральных совокупностей равны между собой, уровень значимости – 5%.
> cochran.test(object= c(var(x1),var(x2),var(x3),var(x4)), data=c(7,9,5,4))
Cochran test for outlying variance
data: c(var(x1), var(x2), var(x3), var(x4))
C = 0.4499, df = 6.25, k = 4.00, p-value = 0.3083
alternative hypothesis: Group 2 has outlying variance
Значения
Cochran C = 0.4499 (значение критериальной статистики теста Кохрана), число степеней свободы (средний объем выборки) 6.25, число групп 4, p -value 0.3083. Альтернативная гипотеза – дисперсия второй выборки значительно больше остальных (является «выбросом»). Поскольку p -value = 0.3083, то отвергнуть гипотезу H0 можно только при допустимой ошибке в 30.83%. Следовательно, гипотеза об однородности дисперсий принимается на 5% уровне значимости.
Дисперсионный анализ
Описание
Данный метод основан на разложении общей дисперсии численного признака на составляющие ее компоненты (отсюда и название метода ANalysis Of VAriance или ANOVA), сравнивая которые с друг другом посредством F‑критерия Фишера можно определить, какую долю (по отношению к совокупности случайных причин) общей вариации признака обуславливает действие на него известных величин (факторов).
Метод основан на сравнении межгрупповой и внутригрупповой изменчивости признака. Каждую группу образуют значения признака при фиксированных значениях (уровнях) известных факторов, поэтому единственным источником дисперсии (изменчивости) внутри каждой группы является суммарное воздействие совокупности случайных причин. Общая модель дисперсионного анализа (на примере двух факторов) выглядит следующим образом:

где  - среднее значение признака,
- среднее значение признака,  - влияние первого фактора на i -м уровне (при i -м значении),
- влияние первого фактора на i -м уровне (при i -м значении),  - влияние второго фактора на j -м уровне (при j -м значении),
- влияние второго фактора на j -м уровне (при j -м значении), 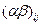 - влияние взаимодействия факторов на указанных уровнях (если факторы не независимы), и
- влияние взаимодействия факторов на указанных уровнях (если факторы не независимы), и 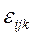 - суммарное влияние на признак случайных факторов, имеющее нормальное распределение с нулевым матожиданием и дисперсией
- суммарное влияние на признак случайных факторов, имеющее нормальное распределение с нулевым матожиданием и дисперсией  . Предполагается, что
. Предполагается, что  не зависит от уровней факторов, поэтому общая дисперсия признака (точнее, общая сумма квадратов
не зависит от уровней факторов, поэтому общая дисперсия признака (точнее, общая сумма квадратов  , где точки в индексе среднего показывают, по каким из них проводилось осреднение, может быть разложена на компоненты (частные суммы), соотвествущие вкладу в общую дисперсию каждой составляющей.
, где точки в индексе среднего показывают, по каким из них проводилось осреднение, может быть разложена на компоненты (частные суммы), соотвествущие вкладу в общую дисперсию каждой составляющей.
В простейшем случае, если имеется всего один фактор, такое разложение представляется в виде таблицы дисперсионного анализа:

| Источник дисперсии | SS сумма квадратов | Степеней свободы | Средний квадрат | F статистика |
| Фактор (межгрупповая) | 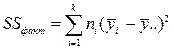
| 
| 
| 
|
| Случйная составляющая | 
| 
| 
| |
| Общая | 
| 
| 
|
где 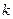 - число групп,
- число групп,  - число наблюдений в i -ой группе,
- число наблюдений в i -ой группе,  - общее число наблюдений.
- общее число наблюдений.
Для проведения однофакторного дисперсионного анализа в R используется линейная модель, в которой единственной независимой переменной выступает этот фактор.
Описание функции
anova(object)
Параметры
| object | Объект класса lm, glm. |
В примере ниже формируется набор данных, включающий 20 значений признака (вектор weight, по 10 значений для каждого из двух уровней фактора и вектор значений фактора group), строится модель зависимости признака от фактора, и выполняется дисперсионный анализ. В случае однофакторной модели таблица дисперсионного анализа совпадает с результатом дисперсионного анализа модели – сравнением остаточной и модельной дисперсий (последняя является суммарным вкладом всех факторов).
Пример
> ctl <- c(4.17,5.58,5.18,6.11,4.50,4.61,5.17,4.53,5.33,5.14)
> trt <- c(4.81,4.17,4.41,3.59,5.87,3.83,6.03,4.89,4.32,4.69)
> group <- gl(2,10,20, labels=c("Ctl","Trt"))
> weight <- c(ctl, trt)
> boxplot(weight ~ group)
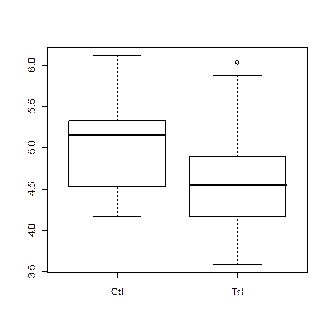
|
> lm.D <- lm(weight ~ group)
> summary(lm.D)
Residuals:
Min 1Q Median 3Q Max
-1.0710 -0.4938 0.0685 0.2462 1.3690
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 4.8465 0.1557 31.124 <2e-16 ***
group1 -0.1855 0.1557 -1.191 0.249
Residual standard error: 0.6964 on 18 degrees of freedom
Multiple R-Squared: 0.07308, Adjusted R-squared: 0.02158
F-statistic: 1.419 on 1 and 18 DF, p-value: 0.249
> anova(lm.D)
Analysis of Variance Table
Response: weight
Df Sum Sq Mean Sq F value Pr(>F)
group 1 0.6882 0.6882 1.4191 0.249 факторная дисперсия
Residuals 18 8.7293 0.4850 внутригрупповая дисперсия
Значения
Как информация по модели summary(), так и вызов anova() выдают для однофакторной модели те же результаты: значение F-статистики 1.419 при 1 и 18 степенях свободы, нулевая гипотеза H0 гласит, что фактор group не влияет на признак weight, уровень значимости (p -value) – 0.249, что означает, что гипотеза может быть отвергнута только если допустить 24.9% ошибки. Таким образом, гипотеза об остуствии вличния фактора принимается на 5% уровне значимости.
Корреляционный анализ